Graduate Student Lefebvre Creates Algorithm to Unlock Mitochondria’s Influence on Diseases

Aug. 23, 2021 – Mitochondria, the organelles inside cells responsible for making energy, hold keys to understanding diseases like cancer, diabetes and Alzheimer’s. Pursuing an interest in studying metastasis – the spread of cancer, Austin Lefebvre, Samueli School graduate student researcher in biomedical engineering, created an algorithm called Mitometer to track mitochondria in live-cell two-dimensional and three-dimensional time-lapse images. His research is published in the August 19, 2021 issue of Nature Methods.
Research over the past decade has shown that mitochondria move to the edges of cancer cells to migrate to other parts of the body. Lefebvre began investigating mitochondrial motility in the Laboratory for Fluorescence Dynamics under Michelle Digman, co-principal investigator and associate professor of biomedical engineering. What he thought would be a simple task – create an algorithm to study the mitochondria’s movement to the edges of breast cancer cells during migration – ended up taking more than a year and a half of iterative work.
“Professor Digman has been super supportive of my ideas,” said Lefebvre. “She’s been encouraging and helping me with any experiments or collaborations that were involved with the project.”
Digman said, “Austin wanted to create our own automated program for mitochondria dynamics that was capable of tracking mitochondria and isolating information in an unbiased way; all the available programs out there didn't have these capabilities. Austin initiated this work and called it ‘Mitometer.’ He worked tirelessly on this project and was able to track mitochondria in organoid tumors that he was imaging with our collaborators.”
Along with Lefebvre and Digman, Dennis Ma and Kai Kessenbrock, department of biological chemistry, and Devon Lawson, physiology and biophysics, also contributed to the research.
Mitochondria are very complex in shape and size, and they vary between cell types and cancers. So making an algorithm to analyze shape, track movement, and work in the general cell biology field proved challenging.
“A lot of people have made algorithms before that are very specific for their microscope or cell type, and mitochondria that are specific sizes, specific shapes,” explained Lefebvre. “But none of them are shape or size independent. That was messing up my progress, so I wanted to make it easier for myself and share it with everyone to let them use it too.”
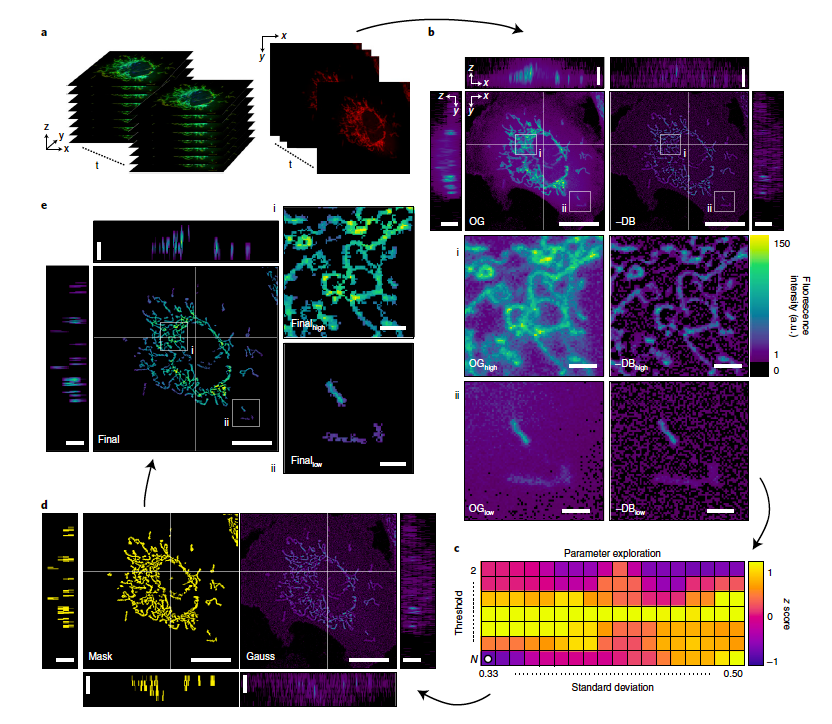
What differentiates Mitometer from other programs is its speed, automation and lack of bias. Mitometer only requires the pixel size and time between frames in the time-lapse images to identify mitochondrial motility and morphology. The segmentation algorithm isolates individual mitochondria by removing the background but preserving the shape and size. The tracking algorithm links mitochondria via differences in morphological features and displacement.
Generally, previous types of tracking and segmenting algorithms are often mostly manual, requiring tracing the shape of the object or clicking individual mitochondria.
“Let’s say you have a video of an object and you have 10 frames,” explained Lefebvre. “You’d be clicking on the same mitochondria for 10 frames that follow it. But naturally, your eye is going to go to those that are maybe clearer or moving quicker or are bigger than the rest. So in that sense, you might be more biased toward whatever you can see, whatever you can recognize.”
Lefebvre’s algorithm doesn’t use any of those factors. It only requires objective features like pixel size, time between each frame and video length. “That removes the bias essentially,” said Lefebvre.
He created a software package of multiple algorithms to make it easier for people to use. Even though the shape of mitochondria differ, the algorithm adapts based on the cell type and other factors. Basically, it’s a self-iterating, self-adapting algorithm that will allow it to work with any time-lapse image, no matter what it looks like.
“The algorithms are completely automated and bias-free to allow people to just put in a time-lapse image of mitochondria in the cell and be able to get back out all of the morphological and motility parameters and see how these mitochondria are splitting apart or coming back together,” said Lefebvre.
In one application using Mitometer, Lefebvre showed that mitochondria of triple-negative breast cancer cells are faster, more directional and more elongated than those in their receptor-positive counterparts. He also found that mitochondrial motility and morphology in breast cancer correlate with metabolic activity, adding key information for developing better targeted treatments.
“The algorithm and software will make studying these mitochondria a lot easier for researchers,” said Lefebvre. “By using the software, it can help identify the mitochondrial differences, so that down the line, we can have better ideas about how to target the more aggressive types of cancer and other diseases like diabetes, Alzheimer’s, Huntington’s and Parkinson’s.”
“Austin is amazingly resourceful, bright and talented,” said Digman. “I put my complete faith in him to be able to lead the project with some guidance. I’m really proud of what he has accomplished!”
– Tonya Becerra
